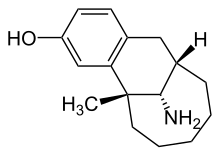
Dezocin je organsko jedinjenje, koje sadrži 16 atoma ugljenika i ima molekulsku masu od 245,360 Da.[6][7]
Dezocin
|
|
|
| Klinički podaci
|
| AHFS/Drugs.com
|
Monografija
|
| Identifikatori
|
| CAS broj
|
53648-55-8
|
| ATC kod
|
N02AX03
|
| PubChem[1][2]
|
3033053
|
| DrugBank
|
DB01209
|
| ChemSpider[3]
|
2297867
|
| KEGG[4]
|
C08010  Y Y
|
| ChEBI
|
CHEBI:4474  Y Y
|
| ChEMBL[5]
|
CHEMBL1685  Y Y
|
| Hemijski podaci
|
| Formula
|
C16H23NO
|
| Mol. masa
|
245,360
|
| SMILES
|
eMolekuli & PubHem
|
| InChI |
|---|
InChI=1S/C16H23NO/c1-16-8-4-2-3-5-12(15(16)17)9-11-6-7-13(18)10-14(11)16/h6-7,10,12,15,18H,2-5,8-9,17H2,1H3/t12-,15-,16+/m0/s1
Key: VTMVHDZWSFQSQP-VBNZEHGJSA-N  Y Y |
|
| Farmakokinetički podaci
|
| Poluvreme eliminacije
|
2,2 h
|
| Farmakoinformacioni podaci
|
| Trudnoća
|
?
|
| Pravni status
|
|
- ↑ Li Q, Cheng T, Wang Y, Bryant SH (2010). „PubChem as a public resource for drug discovery.”. Drug Discov Today 15 (23-24): 1052-7. DOI:10.1016/j.drudis.2010.10.003. PMID 20970519. edit
- ↑ Evan E. Bolton, Yanli Wang, Paul A. Thiessen, Stephen H. Bryant (2008). „Chapter 12 PubChem: Integrated Platform of Small Molecules and Biological Activities”. Annual Reports in Computational Chemistry 4: 217-241. DOI:10.1016/S1574-1400(08)00012-1.
- ↑ Hettne KM, Williams AJ, van Mulligen EM, Kleinjans J, Tkachenko V, Kors JA. (2010). „Automatic vs. manual curation of a multi-source chemical dictionary: the impact on text mining”. J Cheminform 2 (1): 3. DOI:10.1186/1758-2946-2-3. PMID 20331846. edit
- ↑ Joanne Wixon, Douglas Kell (2000). „Website Review: The Kyoto Encyclopedia of Genes and Genomes — KEGG”. Yeast 17 (1): 48–55. DOI:10.1002/(SICI)1097-0061(200004)17:1<48::AID-YEA2>3.0.CO;2-H.
- ↑ Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, Overington JP. (2012). „ChEMBL: a large-scale bioactivity database for drug discovery”. Nucleic Acids Res 40 (Database issue): D1100-7. DOI:10.1093/nar/gkr777. PMID 21948594. edit
- ↑ Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A, Pon A, Banco K, Mak C, Neveu V, Djoumbou Y, Eisner R, Guo AC, Wishart DS (2011). „DrugBank 3.0: a comprehensive resource for omics research on drugs”. Nucleic Acids Res. 39 (Database issue): D1035-41. DOI:10.1093/nar/gkq1126. PMC 3013709. PMID 21059682. edit
- ↑ David S. Wishart, Craig Knox, An Chi Guo, Dean Cheng, Savita Shrivastava, Dan Tzur, Bijaya Gautam, and Murtaza Hassanali (2008). „DrugBank: a knowledgebase for drugs, drug actions and drug targets”. Nucleic Acids Res 36 (Database issue): D901-6. DOI:10.1093/nar/gkm958. PMC 2238889. PMID 18048412. edit
- ↑ Ghose, A.K., Viswanadhan V.N., and Wendoloski, J.J. (1998). „Prediction of Hydrophobic (Lipophilic) Properties of Small Organic Molecules Using Fragment Methods: An Analysis of AlogP and CLogP Methods”. J. Phys. Chem. A 102: 3762-3772. DOI:10.1021/jp980230o.
- ↑ Tetko IV, Tanchuk VY, Kasheva TN, Villa AE. (2001). „Estimation of Aqueous Solubility of Chemical Compounds Using E-State Indices”. Chem Inf. Comput. Sci. 41: 1488-1493. DOI:10.1021/ci000392t. PMID 11749573. edit
- ↑ Ertl P., Rohde B., Selzer P. (2000). „Fast calculation of molecular polar surface area as a sum of fragment based contributions and its application to the prediction of drug transport properties”. J. Med. Chem. 43: 3714-3717. DOI:10.1021/jm000942e. PMID 11020286. edit
